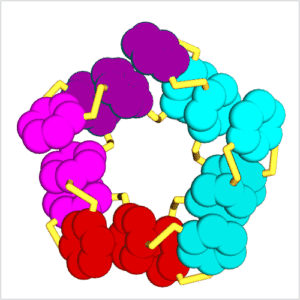
Our research focuses on systems chemistry and explores the interface between supramolecular chemistry, biology, catalysis and materials science. We study complex mixtures of interacting and interconverting molecules with particular focus on the new properties that can emerge from molecules acting collectively. The most fascinating emergent property is life and one of our main aims is to create a completely synthetic form of life. A powerful tool for achieving emergent behaviour is dynamic combinatorial chemistry. We make dynamic combinatorial libraries by connecting small building blocks through reversible covalent bonds to produce a dynamic mixture of molecules that continuously exchange building blocks. When combining reversible covalent bond formation with non-covalent interactions, exciting behaviour can emerge, such as self-replication, the spontaneous formation of surprisingly complex folded molecules, catalytically and even metabolically active assemblies, etc. A brief overview of our work on these phenomena is provided below.
Systems Chemistry[1]
“Systems chemistry is not a subfield, but rather a developmental stage of chemistry”
Dr. Ivica Cvrtila, PhD Thesis, University of Groningen, 2022.
Traditionally chemists have been trained to work with molecules in isolation, i.e., pure compounds. Indeed, for many decades unravelling the structure and determining the properties of pure substances was at the limit of existing capabilities. However, with the recent developments in analytical techniques, it is now possible to study mixtures of molecules and determine their composition and the interactions between them, giving rise to the new field of systems chemistry.[1] What makes this field particularly impactful is that mixtures of interacting molecules can exhibit properties that go well beyond those of pure substances, opening up new horizons in chemistry. These include phenomena that are normally only associated with living systems, such as self-replication, metabolism and even evolution, as we have recently shown.
From Self-Replication to the De-Novo Synthesis of Life[2,3]
In 2010, we reported in Science the spontaneous emergence of two different self-replicating peptide-derived macrocycles from a complex mixture of interconverting molecules.[4] Both replicators compete for an identical feedstock. Competition is influenced by the mode of agitation: shaking favours one replicator while stirring leads to dominance of the other one. The process of self-replication is driven by self-organisation of the replicators into fibres, large enough to be observed by electron microscopy. Replication is exponential as a result of the breakage (division) of growing fibres. In collaboration with the Roos group, we have recently been able to observe replication in real-time by high-speed AFM.[5]
We have since extended this new mechanism of replication to other types of building blocks and have observed exciting phenomena, including the emergence of parasites and even predatory behaviour, where one replicator feeds on another.[6] We also observed stochastic effects (where under identical conditions different replicators may emerge)[7] and behaviour that resembles the process through which different species form in biology.[8]
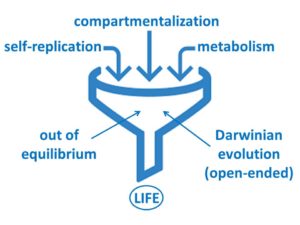
Our main aim is now to extend these systems and create a new and fully synthetic form of life, which has all the main characteristics of life as we know it (replication, metabolism, compartmentalization and the ability to undergo Darwinian evolution), but is built from completely synthetic molecules.
The remaining steps in the synthesis of life which we are currently investigating include: (i) the integration of self-replication with compartmentalization, to create cell-like entities and (ii) the evolution of resulting constructs. Through our work a plausible path to a completely synthetic form of life is starting to be unveiled.
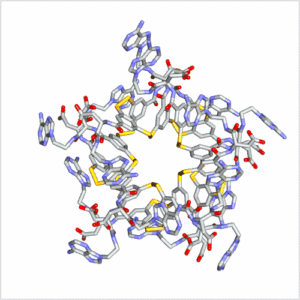
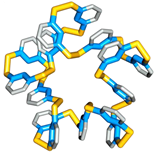 Folding can bestow macromolecules with various properties, as evident from nature’s proteins. Until now complex folded molecules are either the product of evolution or of an elaborate, often iterative, process of design and synthesis. We recently discovered that molecules, that fold in a well-defined architecture of remarkable complexity, can emerge autonomously and selectively from a simple precursor in a single step. We identified self-synthesizing macrocyclic foldamers with complex and unprecedented secondary and tertiary structures. The first example was found to constructs itself highly selectively from 15 identical peptide-nucleobase subunits, using a dynamic combinatorial chemistry approach.[13] Folding of the structure drives its synthesis in 95% yield from a mixture of interconverting molecules of different ring sizes in a one-step process. Single crystal X-ray crystallography and NMR reveals a folding pattern based on an intricate network of noncovalent interactions involving residues space apart widely in the linear sequence. In subsequent work we revealed an entire family of such structures differing in ring size (ranging from 9-23 building blocks) and fold.[14] These results establish dynamic combinatorial chemistry as a powerful approach to developing synthetic molecules with folding motifs of a complexity that goes well beyond that accessible with current design approaches. The fact that such molecules can form autonomously implies that they may have played a role in the emergence life at earlier stages than previously thought possible (i.e. well before the onset of evolution).
Folding can bestow macromolecules with various properties, as evident from nature’s proteins. Until now complex folded molecules are either the product of evolution or of an elaborate, often iterative, process of design and synthesis. We recently discovered that molecules, that fold in a well-defined architecture of remarkable complexity, can emerge autonomously and selectively from a simple precursor in a single step. We identified self-synthesizing macrocyclic foldamers with complex and unprecedented secondary and tertiary structures. The first example was found to constructs itself highly selectively from 15 identical peptide-nucleobase subunits, using a dynamic combinatorial chemistry approach.[13] Folding of the structure drives its synthesis in 95% yield from a mixture of interconverting molecules of different ring sizes in a one-step process. Single crystal X-ray crystallography and NMR reveals a folding pattern based on an intricate network of noncovalent interactions involving residues space apart widely in the linear sequence. In subsequent work we revealed an entire family of such structures differing in ring size (ranging from 9-23 building blocks) and fold.[14] These results establish dynamic combinatorial chemistry as a powerful approach to developing synthetic molecules with folding motifs of a complexity that goes well beyond that accessible with current design approaches. The fact that such molecules can form autonomously implies that they may have played a role in the emergence life at earlier stages than previously thought possible (i.e. well before the onset of evolution).
Click on these images to view an animation of the foldamer crystal structure:
Self-Synthesizing Materials
The autocatalytic self-assembling systems described above also hold considerable promise for materials science. As self-assembly provides the driving force for the formation of very molecules that self-assemble, the resulting materials are self-synthesizing.
The autocatalytic formation of fibres, described above, occurs through a nucleation-growth mechanism. Such mechanism for supramolecular polymer formation resembles living polymerisation and allows controlling fibre length and achieve length distributions with uniquely narrow polydispersity. Our early results are among the first examples of living supramolecular polymerization and allow, for the first time, access to supramolecular block-co-polymers.[15]
We since expanded this work to form hydrogels from these fibres through dynamic covalent crosslinking and have shown, in collaboration with the van Rijn group at the University Medical Center, that these materials are promising platforms for tissue culture.[16]
- R. F. Ludlow, S. Otto
Systems Chemistry.
Chem. Soc. Rev. 2008, 37, 101-108. doi: 10.1039/B611921M. - S. Otto
An Approach to the De Novo Synthesis of Life
Acc. Chem. Res. 2022, 55, 145-155. doi: 10.1021/acs.accounts.1c00534 - P. Adamski, M. Eleveld, A. Sood, Á. Kun, A. Szilágyi, T. Czárán, E. Szathmáry, S. Otto
From Self-Replication to Replicator Systems en Route to De Novo Life
Nature Rev. Chem. 2020, 4, 386–403. doi: 10.1038/s41570-020-0196-x - J. M. A. Carnall, C. A. Waudby, A. M. Belenguer, M. C. A. Stuart, J. J.-P. Peyralans, S. Otto
Mechanosensitive Self-Replication driven by Self-Organization.
Science 2010, 327, 1502-1506. doi: 10.1126/science.1182767. - S. Maity, J. Ottelé, G. Monreal Santiago, P. W. J. M. Frederix, P. Kroon, O. Markovitch, M. C. A. Stuart, S. J. Marrink, S. Otto, W. H. Roos
Caught in the Act: Mechanistic Insight into Supramolecular Polymerization-Driven Self-Replication from Real-Time Visualization
J. Am. Chem. Soc. 2020, 141, 4, 1685-1689. doi: 10.1021/jacs.0c02635 - M. Altay, Y. Altay, S. Otto
Parasitic Behavior of Self‐Replicating Molecules
Angew. Chem. Int. Ed. 2018, 139, 10564–10568. doi:10.1002/anie.201804706. - G. Schaeffer, M. Eleveld, J. Ottelé, P. Kroon, P. Frederix, S. Yang, S. Otto.
Stochastic Emergence of two Distinct Self-Replicators from a Dynamic Combinatorial Library
J. Am. Chem. Soc. 2022, 144, 6291–6297. doi: 10.1021/jacs.1c12591 - J. W. Sadownik, E. Mattia, P. Nowak, S. Otto.
Diversification of Self-Replicating Molecules.
Nature Chem. 2016, 8, 264-269. doi:10.1038/nchem.2419. - G. Monreal Santiago, K. Liu, W. R. Browne, S. Otto
Emergence of Light-Driven Protometabolism on Recruitment of a Photocatalytic Cofactor by a Self-Replicator
Nature Chem. 2020, 12, 603-607. doi: 10.1038/s41557-020-0494-4 - J. Ottelé, A. S. Hussain, C. Mayer, S. Otto
Chance Emergence of Catalytic Activity and Promiscuity in a Self-Replicator
Nature Catal. 2020, 3, 547-553. doi: 10.1038/s41929-020-0463-8 - S. Yang, G. Schaeffer, E. Mattia, O. Markovitch, K. Liu, A. S. Hussain, J. Ottelé, A. Sood, S. Otto
Chemical Fueling Enables Molecular Complexification of Self-Replicators
Angew. Chem. Int. Ed. 2020, 60, 11344-113459. doi: 10.1002/anie.202016196 - Kai Liu, Alex Blokhuis, Chris van Ewijk, Armin Kiani, Juntian Wu, Wouter H. Roos, Sijbren Otto
Light-Driven Eco-Evolutionary Dynamics in a Synthetic Replicator System
Nature Chem. 2023, ASAP. doi: 10.1038/s41557-023-01301-2 - B. Liu, C. G. Pappas, E. Zangrando, N. Demitri, P. J. Chmielewski, S. Otto
Complex Molecules that Fold like Proteins Can Emerge Spontaneously
J. Am. Chem. Soc. 2019, 141, 4, 1685-1689. doi:10.1021/jacs.8b11698. - C. G. Pappas, P. K. Mandal, B. Liu, B. Kauffmann, X. Miao, D. Komáromy, W. Hoffmann, C. Manz, R. Chang, K. Liu, K. Page. I. Huc, S. Otto
Emergence of Low-Symmetry Foldamers from Single Monomers
Nature Chem. 2020, 12, 1180–1186. doi: 10.1038/s41557-020-00565-2 - A. Pal, M. Malakoutikhah, G. Leonetti, M. Tezcan, M. Colomb-Delsuc, V. D. Nguyen, J. van der Gucht, S. Otto.
Controlling the Structure and Length of Self-Synthesizing Supramolecular Polymers through Nucleated Growth and Disassembly.
Angew. Chem. Int. Ed. 2015, 54, 7852–7856. doi: 10.1002/anie.201501965. - I. Marić, L. Yang, X. Li, G. Monreal Santiago, C. G. Pappas, X. Qiu, J. A. Dijksman, K. Mikhailov, P. van Rijn, S. Otto
Tailorable and Biocompatible Supramolecular-Based Hydrogels Featuring two Dynamic Covalent Chemistries
Angew. Chem. Int. Ed. 2023, e202216475. doi: 10.1002/anie.202216475